EPIGENETIC
TARGETS IN THE DIAGNOSIS AND TREATMENT OF PROSTATE CANCER
(
Download pdf )
MURUGESAN MANOHARAN, KAVITHA RAMACHANDRAN, MARK S. SOLOWAY, RAKESH SINGAL
Department of Urology (MM, MSS) and Sylvester Comprehensive and Cancer Center (MM, KR, RS, MSS) Miller School of Medicine, University of Miami, Miami, Florida, USA, and Miami VA Medical Center (RS), Miami, Florida, USA
ABSTRACT
Prostate cancer (PC) is one of leading cause of cancer related deaths in men. Various aspects of cancer epigenetics are rapidly evolving and the role of 2 major epigenetic changes including DNA methylation and histone modifications in prostate cancer is being studied widely. The epigenetic changes are early event in the cancer development and are reversible. Novel epigenetic markers are being studied, which have the potential as sensitive diagnostic and prognostic marker. Variety of drugs targeting epigenetic changes are being studied, which can be effective individually or in combination with other conventional drugs in PC treatment. In this review, we discuss epigenetic changes associated with PC and their potential diagnostic and therapeutic applications including future areas of research.
Key
words: prostate cancer; DNA methylation; epigenesis, genetic
Int Braz J Urol. 2007; 33: 11-8
INTRODUCTION
Prostate
cancer (PC) is one of leading cause of cancer related deaths in men. In
the United States, an estimated 230,000 men were diagnosed with PC in
the year 2005 and approximately 30,000 are expected to die from this cancer
annually (1). Epigenetics refers to stable non inherited changes in the
gene expression without alterations in DNA structure (2).
With the advent of prostate specific antigen
(PSA) in 1998, there has been dramatic increase in the diagnosis of PC
(3). The American Cancer Society recommends annual screening of men above
the age of 50 for PC with PSA and rectal examination (4). However, it
is not clear whether the PSA is effective in the diagnosis of PC as it
lacks both specificity and sensitivity (5,6). About 25% men with normal
PSA may harbor PC (5) and PSA less than 20 ng/mL may not differentiate
between PC and benign conditions (6). This leads to unnecessary prostate
biopsies and on the other hand, we might miss PC in patients with low
PSA. Similarly, there is lack of effective prognostic markers to predict
the behavior of PC and outcome following definitive treatment. Novel biomarkers
based on epigenetic profiling are being explored to aid in the diagnosis
and management of PC (7-9).
Epigenetics is one of the rapidly expanding
fields in cancer related research. Recent studies have shown that epigenetics
plays an important role in cancer biology, somatic gene therapy, viral
infections, genomic imprinting. Epigenetic changes, particularly the DNA
methylation is found to be involved in a variety of cancers including
colon, lung, breast and ovarian cancers apart from prostate cancer (8).
Unlike passively transferred genetic mutations, the epigenetic changes
must be actively maintained and its “reversibility” makes
them a potential therapeutic target (10).
In this review, we discuss epigenetic changes
associated with prostate cancer and their potential diagnostic and therapeutic
applications including future areas of research.
BASICS OF EPIGENETICS
Genome
Approximately 23,000 genes are contained
in the human genome. For proper functioning of the cells, these genes
should be expressed in specific cells at specific times (11). The chromatin
is a nucleoprotein complex made of nucleosomes. The nucleosomes are made
of DNA, which are wrapped around octamers of globular histone proteins
(12). The changes in the chromatin structure influence the gene expression.
When the chromatin is condensed, the gene expression is “switched
off” and when it is open, the gene expression is “switched
on” (13). The status of chromatin is dynamic and can be controlled
by reversible epigenetic mechanisms.
The two important, well studied epigenetic
mechanisms are DNA methylation and histone modifications such as acetylation.
These two processes can act independently and/ or together affecting the
gene expression and in turn the tumorigenesis.
DNA
Methylation in Prostate Cancer
DNA methylation refers to a covalent chemical
modification, resulting in the addition of a methyl (CH3) group at the
C-5 position of the Cytosine ring in the DNA. The human genome is not
uniformly methylated. “CpG islands” are small regions within
the genome that are rich in Cytosine and Guanine bases and are mostly
unmethylated (8). Epigenetic alterations target this region thereby affecting
gene expression. Both the hyper and hypomethylation can affect the gene
expression and the role of DNA methylation in oncogenesis has been studied
for several years.
DNA
Hypermethylation
DNA hypermethylation is a well established
epigenetic abnormality seen in several malignancies, more importantly
in prostate cancer (9). Carcinogenesis is a multi step process and hypermethylation
is hypothesized as an early event in the development and progression of
prostate cancer (14). Hypermethylation of the gene is facilitated by a
group of enzymes known as DNA methyltransferases (DNMT), which includes
DNMT1, DNMT1b, DNMT1o, DNMT1p, DNMT2, DNMT3a, DNMT3b and DNMT3L (15).
The hypermethylation involves the CpG islands in the promoter regions
that results in the silencing of the genes that are involved in tumor
suppressor activity, DNA repair and other critical cellular mechanisms.
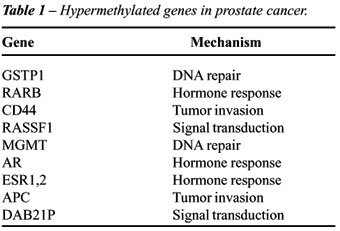
Some of the important genes that are frequently
hypermethylated in prostate cancer are listed in Table-1. Glutathione
S-transferase P1 (GSTP1) is a protector gene and silencing this gene by
hypermethylation leads to DNA damage and cancer initiation (7,16). Methyl
Guanine DNA methyl transferase (MGMT) is another DNA repair gene which
are silenced by hypermethylation (17). Inactivation of putative tumor
suppressor genes by hypermethylation, such as Ras association domain family
1 gene (RASSF1A) (18,19), KAI 1 (20), Inhibin-alpha (21) and DAB21P (22).
Hypermethylation promotes carcinogenesis in prostate cancer by affecting
cell cycle control, hormonal response, cell adhesions and architecture
(14).
DNA Hypomethylation
DNA hypomethylation is a second type of
methylation related epigenetic aberration seen in variety of malignancies
including prostate cancer (23). Hypomethylation is facilitated by enzyme
group demethylases which includes 5-methylcytosine glycosylase and MBD2b
(24). Methylation of normal genomes act as defensive mechanisms against
cancer, for example, the oncogenes can be transcriptionally silenced and
prevented from propogating by being methylated. The hypomethylation causes
breakdown of this defense mechanism and is implicated in the tumor genesis.
The hypomethylation can be “global”
or “localized”. Global hypomethylation refers to overall decrease
in methylation content in the genome. Bedford et al. reported that global
hypomethylation is significantly lower in patients with metastatic prostate
cancer compared to non metastatic prostate cancer (23). Localized or gene
specific hypomethylation refers to a decrease in cytosine methylation
relative to normal levels. This affects the specific regions within genome
such as promoter regions of oncogenes which are highly methylated (9).
Histone
Code
Histones have emerged as important regulators
of chromatin, thereby controlling gene expression. In each nucleosome,
two super helical turns of DNA containing around 146 base pairs wrap an
octomer of histone core made of four histone partners (an H3-H4 tetramer
and two H2A-H2B diamers (25). Histones consist of a globular domain and
a more flexible and charged NH2 terminal called as histone “tail”.
These tails which are placed peripherally are susceptible for a variety
of covalent modifications, such as acetylation, methylation, phosphorylation
and ubiquitination. These modifications are referred as “the histone
code” and is effective epigenetic mechanism regulating gene expression
(26).
Histone acetylation and deacetylations are
mediated by histone acetyl transferases (HAT) and histone deacetylases
(HDAC) respectively. Huang et al. and Tsubaki et al. reported that treatment
of prostate cancer cells with HDAC inhibitors results in increased expression
of specific genes such as CPA3 (27) and Insulin like growth factor binding
protein 3 (28). Coxsackie and adenovirus receptor (CAR) gene and Vitamin
D receptor gene have been shown to be affected by histone acetylation
in prostate cancer. Decreased CAR expression is associated with an increased
Gleason score (29).
Histone methylation affects the chromatin
function depending on the specific amino acid being modified and the extent
of methylation (30). Methylation of H3 at lysine 4 is associated with
inactive transcription of the PSA gene in prostate cancer cell line LNCaP
and decreased di and trimethylated H3 at lysine 4 is associated with AR
mediated transcription of the PSA gene (9). No histone demethylases have
been described so far and it is postulated that histone methylation may
be relatively stable and even irreversible (30).
DNA
Methylation - Histone Code Interplay
DNA promoter methylation and histone deacetylation
can act synergistically resulting in inactive chromatin state resulting
in suppression of gene expression (Figure-1). Methylated DNA binding proteins
such as MeCP2 may play an important role. Retinoic acid receptor beta
gene (RARB) which is silenced in prostate cancer tissues and cell lines
is regulated by both methylation and histone acetylation. This indicates
that combined treatment targeting methylation and histone acetylation
may result in reversal of epigenetic silencing of tumor suppressor genes
(9,31). Similarly, DNA methylation and histone methylation may interact
to facilitate chromatin silencing. However, it is unclear which event
takes place first (9).

Epigenetic Diagnostic Markers
Prostate specific antigen (PSA) is a less
than optimal tumor marker and cannot effectively differentiate between
prostate cancer and other conditions such as prostatitis, benign prostatic
hyperplasia. The false positive results lead to expensive and invasive
investigations such as transrectal prostate biopsy. This provides the
opportunity to the researchers to identify potential epigenetic markers
in the diagnosis of prostate cancer.
Epigenetic markers, particularly aberrant
DNA methylation, have the potential as an useful diagnostic tumor marker.
These markers can be detected in cancer tissues, serum and body fluids.
The methylation markers have several advantages over the mutation based
genetic markers. The detection of these markers is technically simple
and can be sensitively detected both quantitatively and qualitatively
by polymerase chain reaction (PCR). Furthermore, the incidences of aberrant
DNA methylation are higher than those of mutations are and can be discovered
by genome wide screening procedures (32).
In the recent years, the role of GSTP1 is
being studied as a tumor marker widely. Gossel et al. reported that GSTP1
hypermethylation is seen in the serum of 72% patients with of Prostate
cancer patients (33). They also examined the urine after prostatic massage
and methylation was detected in 68% patients with early prostate cancer
and 78% of patients with locally advanced cancer. Table-2 shows methylation
of GSTP1 in different tissue and body fluids. Harden et al. reported 73%
GSTP1 methylation in prostate cancer tissue samples. They also reported
that methylation assay with histological analysis improves the diagnostic
specificity (34). Methylation of several other genes have been studied
in the diagnosis of prostate cancer including, RARB, CD44, E- cadherin
(ECAD), RASSF1A, APC and tazarotene induced gene 1 (T1G1) (7,35). Recent
studies reported by Yegnasubramanian et al. (36) and others have reported
that use of a panel of methylation markers including GSTP1 improves the
diagnosis of prostate cancer both in body fluids and tissues. Further
studies are needed before these markers can be used as diagnostic markers
in the routine clinical practice.

Prognostic
Markers
Kollerman et al. demonstrated that GSTP1
hypermethylation is seen in 40% of pre operative bone marrow aspirate
in patients with advanced PC (37). They also found evidence of GSTP1 hypermethylation
in 90% of PC patients with lymph node involvement where as in only 11%
of lymph nodes in non cancer group. Genes such as CAV1, CDH1, CD 44 and
T1G1 may exhibit specific methylation in high risk and Metastatic tumors
that can be used in the molecular staging and predictors of disease progression
(14). Prostate cancers with high Gleason score are correlated with a higher
degree of methylation of many genes, such as RARβ, RASSF1A, GSTP1
and CDH13 (8). Further studies also indicate that use of panel of multiple
methylation makers can be better predictors than individual genes (38).
THERAPEUTIC TARGETS
Epigenetic
changes are heritable and potentially reversible. Hence, it is reasonable
to expect that these can be used as potential therapeutic targets. Currently
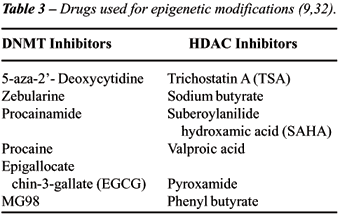
there are several drugs which are at different stages of development.
They can be broadly classified in two groups: (i) DNMT inhibitors and
(ii) Histone Deacetylase (HDAC) inhibitor. Some of the drugs in both groups,
which are being tested and used currently, are shown in Table-3.
DNMT
Inhibitors
5-aza-2’ - Deoxycytidine (5-aza-dC)
is one of the early drugs identified as DNMT inhibitor after being as
cytotoxic drug around 1990. This drug forms irreversible covalent bonds
with DNMT1 after its incorporation in to DNA, thereby inducing degradation
of DNMT1 (39) Issa et al. (40) demonstrated that low dose continuous administration
is more effective than higher doses. Myelosuppression is a known side
effect of this drug which is otherwise well tolerated. 5-aza-dC has been
recently approved by FDA for clinical use in certain hematological conditions.
Another drug in the same group, Zebularine can be administered orally
or intraperitoneally. It has to be given in high doses, however, it is
chemically stable and has low toxicity (41). Other drugs in this which
are being studied include Epigallocatechin-3 - Gallate (EGCG), Procainamide,
Procaine and MG 98 (32).
Histone
Deacetylases (HDAC) Inhibitors
A variety of natural products exhibit HDAC
inhibitory activity. Commonly used HDAC inhibitors which are being tested
include trichostatin A (TSA), Suberoylanilide hydroxamic acid (SAHA) and
valproic acid (9). Many of these drugs have exhibited antitumor activity.
SAHA and sodium butyrate have shown prostate cancer inhibition in animal
models (42,43). Overall, low toxicity rates of these drugs are encouraging
in conducting further studies.
The combination of HDAC and DNMT inhibitors
has synergistic effect in the reactivation of silenced gene (9). Another
interesting possibility is the combination of epigenetic drugs and conventional
anti androgens and chemotherapeutic agents. It should be cautioned that
the epigenetic drugs currently lack gene specificity and some of them
are associated with significant toxicity. Hence, efforts are being made
to develop gene specific epigenetic drugs (32).
SUMMARY AND FUTURE DIRECTIONS
Epigenetic changes in prostate cancer are being studied extensively at present and genome wide screening will lead to development of novel epigenetic markers. Epigenetic changes are early event in cancer development and hence can be used to assess the risk of developing cancer. Li et al. suggest that genes such as CAV1, CDH1, CD44 and T1G1 should be explored further as “risk markers”, particularly to differentiate the indolent tumors from others with bad prognostic potential (14). Epigenetic molecular classification will help to identify patients at high risk of recurrence following definitive treatments such as radical prostatectomy. Therapeutic drugs which reverse these epigenetic changes have the potential to be an effective adjunct treatment for prostate cancer. However, they need to be studied both for its efficacy and safety profile. Gene specific epigenetic drugs need to be developed for better targeting of the disease. As the epigenetic changes are early event in the PC development, these drugs have a potential to play a role in disease prevention. Two main features of epigenetic changes, “reversibility” and being an “early event” in tumorigenesis, makes epigenetic targeting as an important future research area for cancer diagnosis, risk stratification, treatment and prevention resulting in effective cancer control.
CONFLICT OF INTEREST
None declared.
REFERENCES
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, et al.: Cancer statistics, 2005. CA Cancer J Clin. 2005; 55: 10-30. Erratum in: CA Cancer J Clin. 2005; 55: 259.
- Wolffe AP, Matzke MA: Epigenetics: regulation through repression. Science. 1999; 286: 481-6.
- Crawford ED: Epidemiology of prostate cancer. Urology. 2003; 62: 3-12.
- Smith RA, Cokkinides V, Eyre HJ: American Cancer Society guidelines for the early detection of cancer, 2003. CA Cancer J Clin. 2003; 53: 27-43.
- Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, et al.: Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004; 350: 2239-46. Erratum in: N Engl J Med. 2004; 351: 1470.
- Stamey TA, Caldwell M, McNeal JE, Nolley R, Hemenez M, Downs J: The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol. 2004; 172: 1297-301.
- Singal R, Ferdinand L, Reis IM, Schlesselman JJ: Methylation of multiple genes in prostate cancer and the relationship with clinicopathological features of disease. Oncol Rep. 2004; 12: 631-7.
- Das PM, Singal R: DNA methylation and cancer. J Clin Oncol. 2004; 22: 4632-42.
- Li LC, Carroll PR, Dahiya R: Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J Natl Cancer Inst. 2005; 97: 103-15.
- Egger G, Liang G, Aparicio A, Jones PA: Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004; 429: 457-63.
- Rodenhiser D, Mann M: Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 2006; 174: 341-8.
- Peterson CL, Laniel MA: Histones and histone modifications. Curr Biol. 2004; 14: R546-51.
- Rountree MR, Bachman KE, Herman JG, Baylin SB: DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001; 20: 3156-65.
- Li LC, Okino ST, Dahiya R: DNA methylation in prostate cancer. Biochim Biophys Acta. 2004; 1704: 87-102.
- Robertson KD: DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002; 21: 5361-79.
- Nelson CP, Kidd LC, Sauvageot J, Isaacs WB, De Marzo AM, Groopman JD, et al.: Protection against 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine cytotoxicity and DNA adduct formation in human prostate by glutathione S-transferase P1. Cancer Res. 2001; 61: 103-9.
- Konishi N, Nakamura M, Kishi M, Nishimine M, Ishida E, Shimada K: DNA hypermethylation status of multiple genes in prostate adenocarcinomas. Jpn J Cancer Res. 2002; 93: 767-73.
- Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, et al.: Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006; 66: 2562-9.
- Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, et al.: The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004; 6: 129-37.
- Sekita N, Suzuki H, Ichikawa T, Kito H, Akakura K, Igarashi T, et al.: Epigenetic regulation of the KAI1 metastasis suppressor gene in human prostate cancer cell lines. Jpn J Cancer Res. 2001; 92: 947-51.
- Schmitt JF, Millar DS, Pedersen JS, Clark SL, Venter DJ, Frydenberg M, et al.: Hypermethylation of the inhibin alpha-subunit gene in prostate carcinoma. Mol Endocrinol. 2002; 16: 213-20.
- Chen H, Toyooka S, Gazdar AF, Hsieh JT: Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J Biol Chem. 2003; 278: 3121-30.
- Bedford MT, van Helden PD: Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987; 47: 5274-6.
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M: A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999; 397: 579-83.
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389: 251-60.
- Jenuwein T, Allis CD: Translating the histone code. Science. 2001; 293: 1074-80.
- Huang H, Reed CP, Zhang JS, Shridhar V, Wang L, Smith DI: Carboxypeptidase A3 (CPA3): a novel gene highly induced by histone deacetylase inhibitors during differentiation of prostate epithelial cancer cells. Cancer Res. 1999; 59: 2981-8.
- Tsubaki J, Hwa V, Twigg SM, Rosenfeld RG: Differential activation of the IGF binding protein-3 promoter by butyrate in prostate cancer cells. Endocrinology. 2002; 143: 1778-88.
- Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V, et al.: Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: potential relevance to gene therapy. Cancer Res. 2002; 62: 3812-8.
- Moggs JG, Goodman JI, Trosko JE, Roberts RA: Epigenetics and cancer: implications for drug discovery and safety assessment. Toxicol Appl Pharmacol. 2004; 196: 422-30.
- Nakayama T, Watanabe M, Yamanaka M, Hirokawa Y, Suzuki H, Ito H, et al.: The role of epigenetic modifications in retinoic acid receptor beta2 gene expression in human prostate cancers. Lab Invest. 2001; 81: 1049-57.
- Miyamoto K, Ushijima T: Diagnostic and therapeutic applications of epigenetics. Jpn J Clin Oncol. 2005; 35: 293-301.
- Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, et al.: Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000; 60: 5941-5.
- Harden SV, Guo Z, Epstein JI, Sidransky D: Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J Urol. 2003; 169: 1138-42.
- Tokumaru Y, Harden SV, Sun DI, Yamashita K, Epstein JI, Sidransky D: Optimal use of a panel of methylation markers with GSTP1 hypermethylation in the diagnosis of prostate adenocarcinoma. Clin Cancer Res. 2004; 10: 5518-22.
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, et al.: Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004; 64: 1975-86.
- Kollermann J, Muller M, Goessl C, Krause H, Helpap B, Pantel K, et al.: Methylation-specific PCR for DNA-based detection of occult tumor cells in lymph nodes of prostate cancer patients. Eur Urol. 2003; 44: 533-8.
- Rhodes DR, Sanda MG, Otte AP, Chinnaiyan AM, Rubin MA: Multiplex biomarker approach for determining risk of prostate-specific antigen-defined recurrence of prostate cancer. J Natl Cancer Inst. 2003; 95: 661-8.
- Christman JK: 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002; 21: 5483-95.
- Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, et al.: Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004; 103: 1635-40.
- Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, et al.: Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004; 24: 1270-8.
- Butler LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, et al.: Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000; 60: 5165-70.
- Kuefer R, Hofer MD, Altug V, Zorn C, Genze F, Kunzi-Rapp K, et al.: Sodium butyrate and tributyrin induce in vivo growth inhibition and apoptosis in human prostate cancer. Br J Cancer. 2004; 90: 535-41.
________
Accepted:
September 9, 2006
_______________________
Correspondence address:
Dr. Murugesan Manoharan
Associate Professor of Urology
University of Miami School of Medicine
P.O. Box 016960 (M814)
Miami, Florida, 33101, USA
Fax: + 1 305 243-4653
E-mail: mmanoharan@med.miami.edu